Matching patterns
2023-03-10
Introduction
The problem
What’s different between these data sets?
What is needed to create data2 from data1?
data1# A tibble: 12 × 3
time species resp
<chr> <chr> <chr>
1 early-day dogfish yes
2 mid-day bear dog no
3 late-day dog yes
4 daytime dogfish no
5 early-day cat yes
6 mid-day cat no
7 late-day dogfish no
8 daytime bear dog no
9 early-day dogfish <NA>
10 mid-day catfish yes
11 late-day cat yes
12 daytime bear dog yes data2# A tibble: 8 × 3
time species resp
<chr> <chr> <chr>
1 early-Day dogfish yes
2 mid-Day bear dog no
3 late-Day dog yes
4 daytime dogfish no
5 late-Day dogfish no
6 daytime bear dog no
7 early-Day dogfish no data
8 daytime bear dog yes Set-up
Mental model
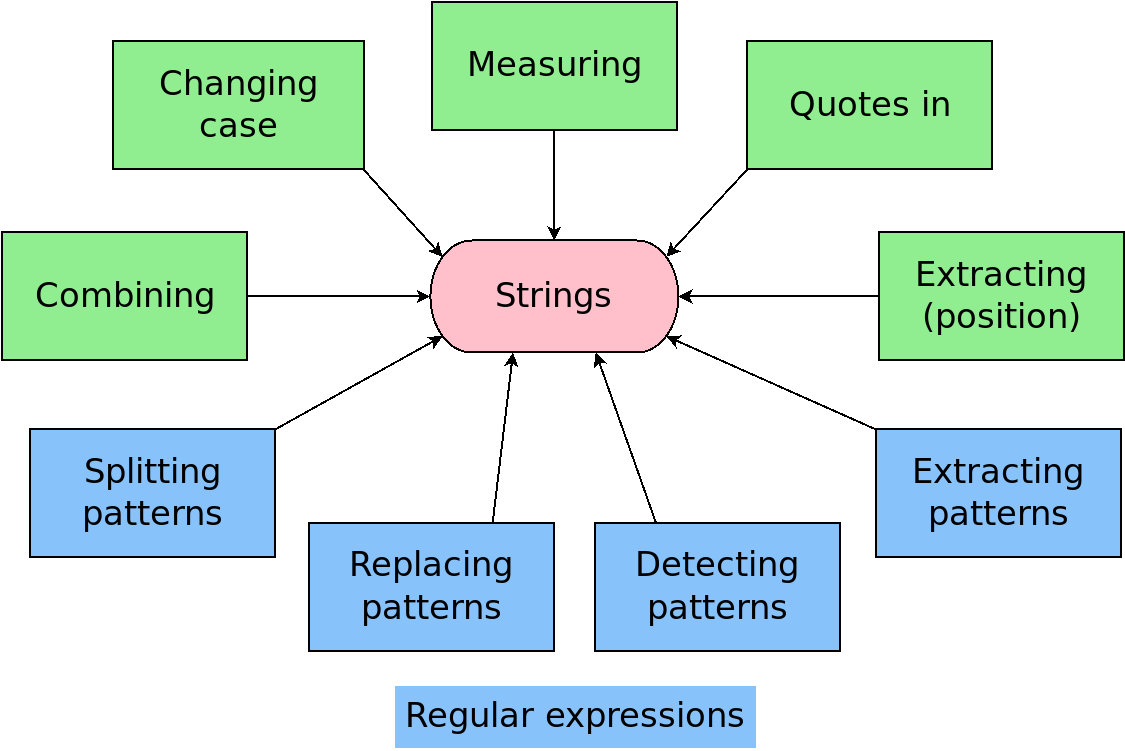
Strings with {stringr}


Patterns
Regular expressions
Concise and powerful language for describing patterns within strings
(regex for short)

Regular expressions
Here’s the regex I used to detect IP addresses: ^(?:(25[0-5]|2[0-4][0-9]|1[0-9][0-9]|[1-9][0-9]|[0-9])(\.(?!$)|$)){4}$

Matching strings
View string patterns with stringr::str_view_all()
(x <- c("apple", "banana", "pear", NA))[1] "apple" "banana" "pear" NA str_view_all(x, "a")[1] │ <a>pple
[2] │ b<a>n<a>n<a>
[3] │ pe<a>r
[4] │ NARegex 101
. is wildcard
str_view_all(x, ".a.")[1] │ apple
[2] │ <ban>ana
[3] │ p<ear>
[4] │ NARegex 101
^ to match the start of the string (like starts_with())
$ to match the end of the string (like ends_with())
str_view_all(x, "^a")[1] │ <a>pple
[2] │ banana
[3] │ pear
[4] │ NAstr_view_all(x, "a$")[1] │ apple
[2] │ banan<a>
[3] │ pear
[4] │ NARegex 101
| matches one pattern OR another (e.g., this|that)
str_view_all(x, "ap|an|ar")[1] │ <ap>ple
[2] │ b<an><an>a
[3] │ pe<ar>
[4] │ NAWrap character groups in ()
str_view_all("Are you here or are you there?", "(A|a)re") [1] │ <Are> you here or <are> you there?Regex 101
\d matches any digit
# view digits
str_view_all("March 10, 2020", "\\d")[1] │ March <1><0>, <2><0><2><0>Regex 101
[abc] matches individual characters (a, b, or c)
# view everything with ab or a<space>
str_view_all(c("abc", "a.c", "a*c", "a c"), "a[b ]")[1] │ <ab>c
[2] │ a.c
[3] │ a*c
[4] │ <a >cRegex 101
[^abc] matches individual characters except a, b, or c
# view everything except ab and a<space>
str_view_all(c("abc", "a.c", "a*c", "a c"), "a[^b ]")[1] │ abc
[2] │ <a.>c
[3] │ <a*>c
[4] │ a c# view everything except digits
str_view_all("March 10, 2020", "[^\\d]") [1] │ <M><a><r><c><h>< >10<,>< >2020Detecting and extracting patterns
Detecting pattern matches
Detect matching elements with stringr::str_detect()
x[1] "apple" "banana" "pear" NA str_detect(x, "e") # results in logical vector[1] TRUE FALSE TRUE NAsum(str_detect(x, "e"), na.rm = TRUE) # sum matching elements[1] 2mean(str_detect(x, "e"), na.rm = TRUE) # calculate proportion of matches[1] 0.6666667Extracting pattern matches
Extract observations matching pattern with filter() and str_detect()
penguins |>
filter(str_detect(sex, "male")) |> # select observations that include "male"
select(species, island, sex)# A tibble: 333 × 3
species island sex
<fct> <fct> <fct>
1 Adelie Torgersen male
2 Adelie Torgersen female
3 Adelie Torgersen female
4 Adelie Torgersen female
5 Adelie Torgersen male
6 Adelie Torgersen female
7 Adelie Torgersen male
8 Adelie Torgersen female
9 Adelie Torgersen male
10 Adelie Torgersen male
# … with 323 more rowsExtracting pattern matches
Extract elements that match a pattern with stringr::str_subset()
head(words, n = 20) [1] "a" "able" "about" "absolute" "accept" "account"
[7] "achieve" "across" "act" "active" "actual" "add"
[13] "address" "admit" "advertise" "affect" "afford" "after"
[19] "afternoon" "again" str_subset(words, "^rec") # select elements starting with "rec"[1] "receive" "recent" "reckon" "recognize" "recommend" "record" str_subset(words, "ing$") # select elements ending with "ing"[1] "bring" "during" "evening" "king" "meaning" "morning" "ring"
[8] "sing" "thing" Replacing patterns
Replacing pattern matches
Replace matches with new strings with stringr::str_replace() and stringr::str_replace_all()
str_replace(x, "[aeiou]", "-") # replace only first instance of match[1] "-pple" "b-nana" "p-ar" NA str_replace_all(x, "[aeiou]", "-") # replace all matches[1] "-ppl-" "b-n-n-" "p--r" NA str_replace_all(x, "[^aeiou]", "-") # replace all matches[1] "a---e" "-a-a-a" "-ea-" NA Replacing pattern matches
You can use this to recode character variables, but…
set.seed(50)
penguins |>
mutate(new_island = str_replace(island, "Torgersen", "Party")) |>
select(species, island, new_island) |>
slice_sample(n = 6)# A tibble: 6 × 3
species island new_island
<fct> <fct> <chr>
1 Adelie Torgersen Party
2 Chinstrap Dream Dream
3 Adelie Dream Dream
4 Chinstrap Dream Dream
5 Gentoo Biscoe Biscoe
6 Gentoo Biscoe Biscoe It coerces to character data types
I use this A LOT to clean up text data
Replacing NA
Replace NA with another value with stringr::str_replace_na()
x[1] "apple" "banana" "pear" NA str_replace_na(x) # by default replaces NA with "NA"[1] "apple" "banana" "pear" "NA" str_replace_na(x, "Missing") # but you can replace with other strings[1] "apple" "banana" "pear" "Missing"Splitting strings
Splitting strings
Split a string up into pieces with str_split()
head(sentences, n = 2)[1] "The birch canoe slid on the smooth planks."
[2] "Glue the sheet to the dark blue background."[[1]]
[1] "The" "birch" "canoe" "slid" "on" "the" "smooth"
[8] "planks."
[[2]]
[1] "Glue" "the" "sheet" "to" "the"
[6] "dark" "blue" "background."Notice this produces a list. Why?
Splitting strings
Convert to matrix with simplify
Solving the problem
data1# A tibble: 12 × 3
time species resp
<chr> <chr> <chr>
1 early-day dogfish yes
2 mid-day bear dog no
3 late-day dog yes
4 daytime dogfish no
5 early-day cat yes
6 mid-day cat no
7 late-day dogfish no
8 daytime bear dog no
9 early-day dogfish <NA>
10 mid-day catfish yes
11 late-day cat yes
12 daytime bear dog yes data2# A tibble: 8 × 3
time species resp
<chr> <chr> <chr>
1 early-Day dogfish yes
2 mid-Day bear dog no
3 late-Day dog yes
4 daytime dogfish no
5 late-Day dogfish no
6 daytime bear dog no
7 early-Day dogfish no data
8 daytime bear dog yes